What we do...
- Details
Our lab studies cell signaling pathways in gene regulation and cancer. Recent work from the lab includes:
Novel regulators of muscle differentiation
- Details

Gene expression is regulated by functional elements in the genome such as enhancer elements. Tissue-specific gene expression through regulated use of specific enhancer sequences is a major mechanism of development in an organism. Genomics provides powerful tools to study the DNA sequences involved in these processes. We use unbiased proteomics approaches to study proteins and assembled protein machinery at regulatory genomics elements, using a well studied in vitro model of C2C12 muscle differentiation.
We apply DNA baits to pulldown proteins and complexes from nuclear lysates. Using this approach, we identified nearly 20 novel regulators and validated these using shRNA.
Proteomics of cell signaling
- Details
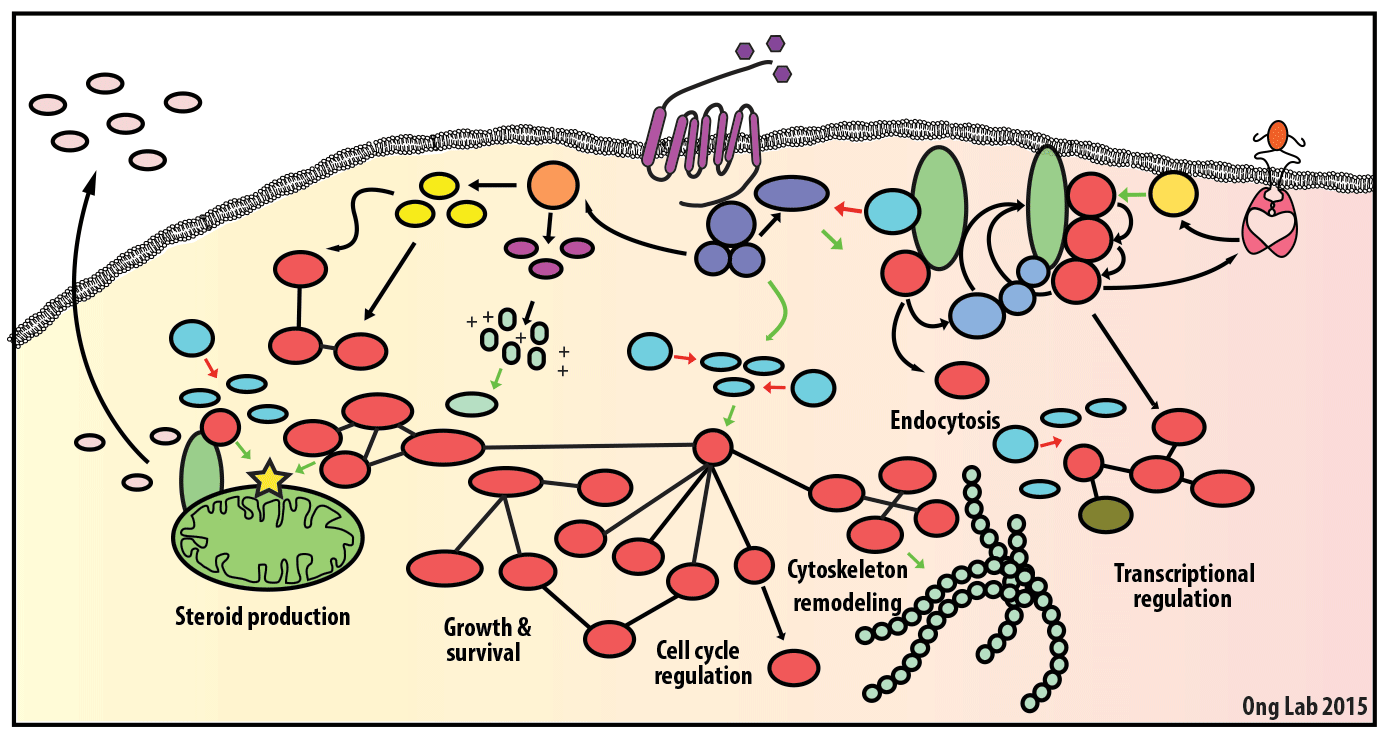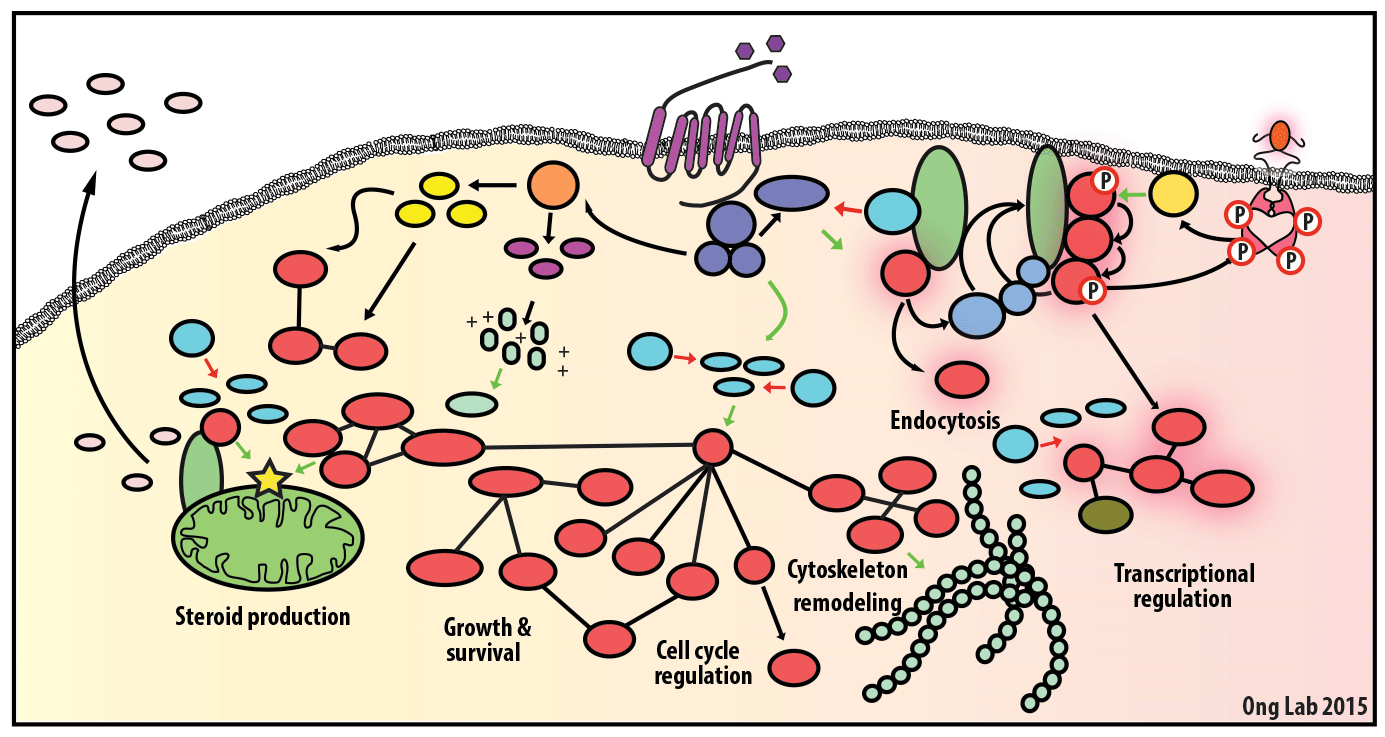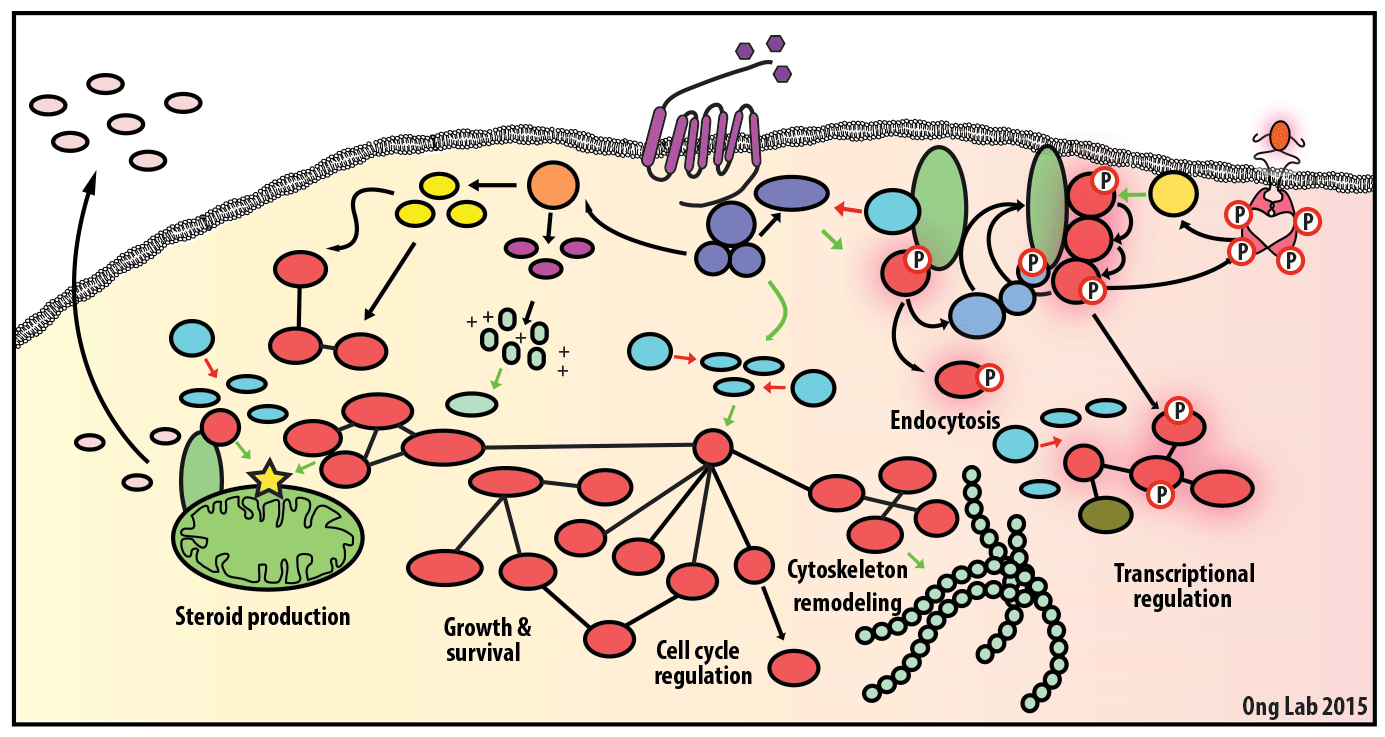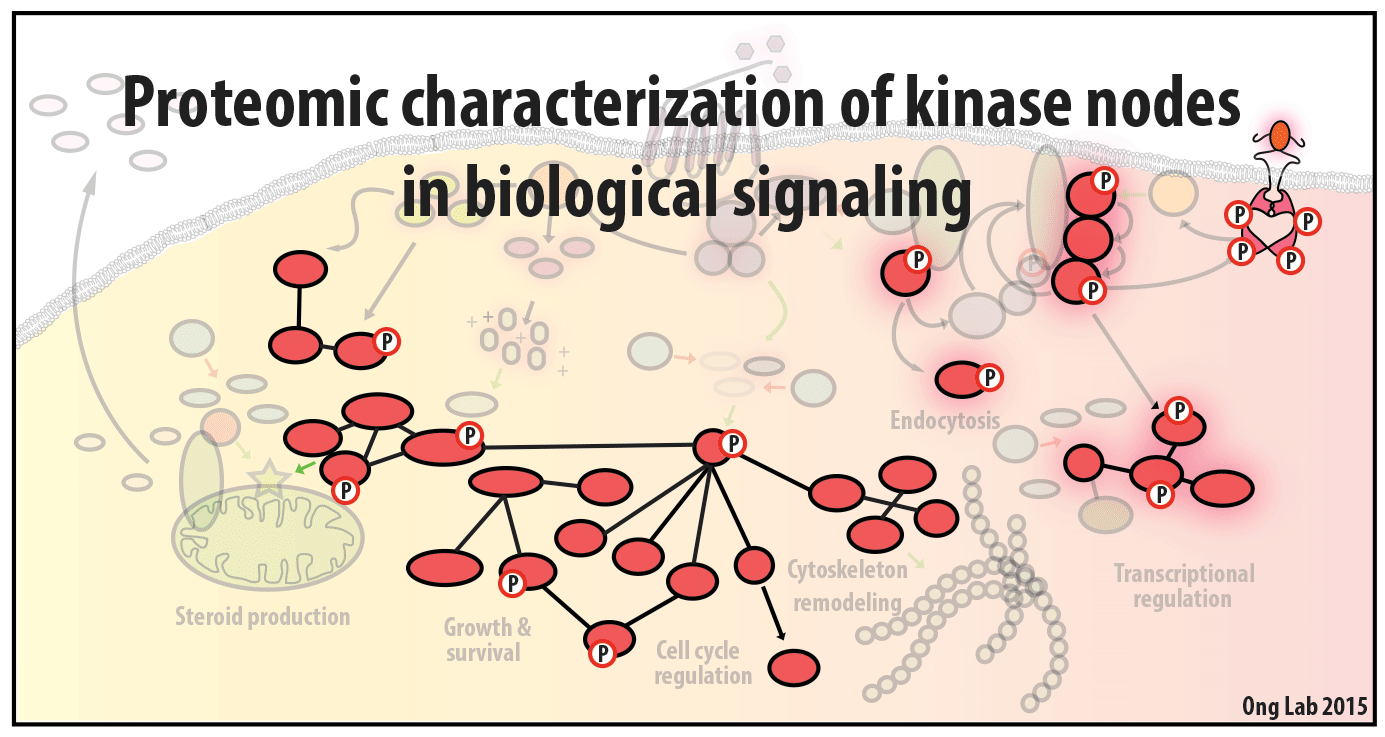
We use mass spectrometry-based analyses to study cell signaling. We have focused on the analyses of kinases, key regulatory nodes in many cellular signaling pathways. To enhance our ability to study these kinases, we use synthesize our own affinity reagents1 that use immobilized non-selective kinase inhibitors to selectively enrich ~70% of the kinases known in the human genome (these affinity reagents are also known as Kinobeads2 or mixed inhibitor beads, MIBs3). This allows us to compare kinase expression and activity between different cellular states, using quantitative proteomics approaches like SILAC.
View our Selected publications for a curated list of work from the lab.
References:
1. Golkowski M, Vidadala RS, Lombard CK, Suh HW, Maly DJ, Ong SE. Kinobead and Single-Shot LC-MS Profiling Identifies Selective PKD Inhibitors. J Proteome Res. 2017;16(3):1216-27. doi: 10.1021/acs.jproteome.6b00817. PubMed PMID: 28102076.
2. Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25(9):1035-44. doi: 10.1038/nbt1328. PubMed PMID: 17721511.
3. Cooper MJ, Cox NJ, Zimmerman EI, Dewar BJ, Duncan JS, Whittle MC, Nguyen TA, Jones LS, Ghose Roy S, Smalley DM, Kuan PF, Richards KL, Christopherson RI, Jin J, Frye SV, Johnson GL, Baldwin AS, Graves LM. Application of multiplexed kinase inhibitor beads to study kinome adaptations in drug-resistant leukemia. PLoS One. 2013;8(6):e66755. doi: 10.1371/journal.pone.0066755. PubMed PMID: 23826126; PMCID: PMC3691232.
Pharmacoproteomics of drug response in cancer
- Details

We developed a new pharmacoproteomics platform combining kinome activity profiling with a kinase inhibitor screen to identify pathway biomarkers of drug response in cancer cell lines.
Correlating 13935 kinome features to response for 299 kinase inhibitors, we scored individual features and 327 Reactome pathways for their relevance to drug action in 17 liver cancer cell lines. This is a great resource for deconvoluting target and mechanisms of action for kinase inhibitors. We developed a Shiny Web app for interactive visualization of this resource: https://quantbiology.org/hcckinome
Our data implicated the EMT in drug resistance and identified novel kinases important for distinct EMT states. Modulating these kinases sensitized resistant cells to drugs, validating our pharmacoproteomic platform to identify rational drug combinations.
With our kinome profiling approach, we can distinguish clinical HCC samples by etiology and identify pathway-based biomarkers in individual patient tumors. Important milestones in developing personalized treatments for HCC.